Determination of the risk of breast and stomach cancer by identifying variants of the 5mC/T polymorphism in the intron of the KIF26B gene at position xp1: 245618129
Akishev A.G., Netesova N.A., Abdurashitov M.A., Vihlyanov I.V., Nikitin M.K., Karpov A.B., Degtyarev S.Kh.
A.G. Akishev1*, N.A.Netesova2, M.A. Abdurashitov1, I.V. Vihlyanov3, M.K. Nikitin3, A.B. Karpov4, S.Kh. Degtyarev1
1 SibEnzyme, Novosibirsk
2 Epigenlab, Novosibirsk
3Altai Regional Oncology Center, 656049, Altai region, Barnaul
4Seversk Biophysical Research Centre, Seversk, Tomsk region
* corresponding author: Alexander Akishev, SibEnzyme, 2/12 Ak.Timakov Str., Novosibirsk 630117, Russia; Tel: +7 383 3334991; Fax: +7 383 3336853; E-mail: aki@sibenzyme.ru
We used GlaI/FatI-PCR assay for determination of 5mC/T polymorphism frequencies at position xp1: 245618129 (according to the GRCh38.p13 genomic assembly) in DNA preparations isolated from blood cells of 51 breast cancer (BC) patients and 63 gastric cancer patients (GC). The data obtained were compared with those previously obtained for 92 healthy blood donors.
It was shown that the cytosine in this position is predominantly is methylated in all DNA samples. The incidence of a diploid C/C set in this position in patients with breast cancer and gastric cancer is 9.8% and 7.94%, respectively. This is significantly different from the incidence of C homozygotes in healthy women (26.09%) and healthy donors of both sexes (22.83%). At the same time, the incidence of C/T heterozygotes and T/T homozygotes in patients with gastric cancer and breast cancer exceeds similar indicators for control groups of healthy people by 5.5-10.7%. Thus, the substitution of a methylated cytosine base for a thymine base at position xp1: 245618129 may be an unfavorable sign indicating a genetic predisposition to two types of oncological diseases.
Keywords: single nucleotide polymorphism 5mC/T, human blood DNA, GlaI-PCR assay, FatI-PCR assay, breast cancer, gastric cancer
DOI: 10.26213/SE.2019.78.40307
Citation:
Akishev A.G., Netesova N.A., Abdurashitov M.A., Vihlyanov I.V., Nikitin M.K., Karpov A.B., Degtyarev S.Kh. (2021) Determination of the risk of breast and stomach cancer by identifying variants of the 5mC/T polymorphism in the intron of the KIF26B gene at position xp1: 245618129, Epigenet DNA diagnx, vol.2021(1), DOI: 10.26213/SE.2019.78.40307
This work is available at Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International
Full-text-pdf
This article is available only on Russian
Table 1. PCR assay data for women with diagnosed breast cancer
Cq values for the cases of hydrolysis of GGCC site (HaeIII), CATG site (Fat-PCR assay) and A(5mC)GC site (GlaI-PCR assay) and difference between Cq values of last two reactions with the first one (ΔCq_T = ΔCq_CATG – ΔCq_GGCC and ΔCq_5mC = ΔCq_A(5mC)GC – ΔCq_GGCC).
|
Samle |
Age |
I Cq_GGCC |
II Cq_CATG |
III Cq_A(5mC)GC |
ΔCq_T (II – I) |
ΔCq_5mC (III-I) |
|
BC57 |
54 |
21.82±0.061 |
22.57±0.208 |
22.6±0.03 |
0.78 |
0.75 |
|
BC58 |
59 |
21.79±0.153 |
21.77±0.149 |
29.35±0.578 |
7.56 |
-0.02 |
|
BC59 |
62 |
22.16±0.055 |
25.34±0.094 |
22.12±0.084 |
-0.04 |
3.18 |
|
BC60 |
60 |
23.22±0.093 |
23.39±0.228 |
28.98±0.139 |
5.76 |
0.17 |
|
BC67 |
36 |
21.56±0.158 |
22.33±0.205 |
22.34±0.094 |
0.78 |
0.77 |
|
BC72 |
65 |
22.41±0.134 |
23.22±0.173 |
23.39±0.204 |
0.98 |
0.81 |
|
BC107 |
50 |
22.99±0.155 |
22.86±0.048 |
31.59±0.487 |
8.6 |
-0.13 |
|
BC117 |
65 |
21.69±0.024 |
21.66±0.032 |
27.97±0.123 |
6.28 |
-0.03 |
|
BC190 |
70 |
21.99±0.023 |
22±0.138 |
29.61±0.522 |
7.62 |
0.01 |
|
BC191 |
62 |
20.58±0.051 |
21.49±0.051 |
21.53±0.025 |
0.95 |
0.91 |
|
BC192 |
66 |
21.57±0.089 |
21.59±0.081 |
29.17±0.511 |
7.6 |
0.02 |
|
BC193 |
69 |
21.75±0.121 |
21.86±0.153 |
28.45±0.085 |
6.7 |
0.11 |
|
BC194 |
65 |
21.31±0.083 |
22.12±0.183 |
22.28±0.099 |
0.97 |
0.81 |
|
BC196 |
73 |
22.31±0.041 |
24.59±0.089 |
22.21±0.023 |
-0.1 |
2.28 |
|
BC197 |
65 |
21.55±0.106 |
22.56±0.109 |
22.54±0.121 |
0.99 |
1.01 |
|
BC198 |
58 |
21.18±0.215 |
21.96±0.169 |
22.29±0.098 |
1.11 |
0.78 |
|
BC199 |
58 |
21.46±0.121 |
21.49±0.078 |
29.71±0.376 |
8.25 |
0.03 |
|
BC201 |
65 |
21.62±0.064 |
21.48±0.087 |
28.81±0.388 |
7.19 |
-0.14 |
|
BC202 |
50 |
21.68±0.09 |
21.68±0.143 |
30.19±0.81 |
8.51 |
0 |
|
BC203 |
78 |
22.24±0.115 |
22.3±0.074 |
30.8±0.582 |
8.56 |
0.06 |
|
BC204 |
31 |
21.67±0.062 |
22.5±0.067 |
22.75±0.036 |
1.08 |
0.83 |
|
BC206 |
50 |
21.4±0.125 |
22.23±0.165 |
22.25±0.137 |
0.85 |
0.83 |
|
BC207 |
63 |
21.52±0.096 |
21.4±0.071 |
29.69±0.518 |
8.17 |
-0.12 |
|
BC208 |
67 |
21.6±0.047 |
22.46±0.043 |
22.58±0.053 |
0.98 |
0.86 |
|
BC292 |
62 |
21.98±0.207 |
23.02±0.005 |
23.17±0.064 |
1.19 |
1.04 |
|
BC293 |
62 |
22.35±0.105 |
22.32±0.007 |
28.77±0.292 |
6.42 |
-0.03 |
|
BC294 |
71 |
22.12±0.028 |
23.1±0.106 |
23.16±0.098 |
1.04 |
0.98 |
|
BC295 |
62 |
22.32±0.036 |
22.11±0.222 |
30.97±0.64 |
8.65 |
-0.21 |
|
BC296 |
58 |
21.3±0.068 |
22.04±0.024 |
22.13±0.093 |
0.83 |
0.74 |
|
BC297 |
70 |
20.7±0.092 |
20.57±0.051 |
28.56±0.617 |
7.86 |
-0.13 |
|
BC298 |
58 |
21.54±0.103 |
21.4±0.098 |
28.92±0.289 |
7.38 |
-0.14 |
|
BC299 |
64 |
21.71±0.021 |
21.55±0.048 |
30.02±0.263 |
8.31 |
-0.16 |
|
BC300 |
66 |
22.1±0.126 |
23.11±0.054 |
22.95±0.028 |
0.85 |
1.01 |
|
BC301 |
44 |
21.63±0.18 |
22.44±0.113 |
22.74±0.125 |
1.11 |
0.81 |
|
BC302 |
63 |
21.71±0.089 |
22.68±0.196 |
22.84±0.04 |
1.13 |
0.97 |
|
BC303 |
58 |
21.71±0.113 |
25.06±0.103 |
21.57±0.066 |
-0.14 |
3.35 |
|
BC304 |
41 |
21±0.069 |
21.58±0.117 |
21.85±0.196 |
0.85 |
0.58 |
|
BC320 |
50 |
23.24±0.045 |
24.24±0.148 |
24.28±0.084 |
1.04 |
1 |
|
BC321 |
48 |
23.22±0.04 |
23.09±0.082 |
29.61±0.692 |
6.39 |
-0.13 |
|
BC322 |
48 |
21.59±0.203 |
22.45±0.188 |
22.46±0.166 |
0.87 |
0.86 |
|
BC323 |
63 |
22.44±0.086 |
22.28±0.037 |
28.35±0.173 |
5.91 |
-0.16 |
|
BC324 |
63 |
22.3±0.092 |
23.06±0.035 |
23.4±0.098 |
1.1 |
0.76 |
|
BC325 |
42 |
21.39±0.109 |
22.3±0.021 |
22.27±0.056 |
0.88 |
0.91 |
|
BC326 |
69 |
22.11±0.019 |
22.18±0.167 |
28.66±0.383 |
6.55 |
0.07 |
|
BC327 |
62 |
22.67±0.096 |
25.64±0.096 |
22.44±0.09 |
-0.23 |
2.97 |
|
BC328 |
44 |
22.38±0.052 |
22.28±0.075 |
29.05±1.272 |
6.67 |
-0.1 |
|
BC329 |
61 |
22.17±0.079 |
23.03±0.048 |
23.06±0.013 |
0.89 |
0.86 |
|
BC330 |
71 |
22.06±0.066 |
22.69±0.119 |
22.99±0.108 |
0.93 |
0.63 |
|
BC331 |
67 |
21.63±0.118 |
22.48±0.048 |
22.58±0.046 |
0.95 |
0.85 |
|
BC332 |
63 |
21.29±0.162 |
22.09±0.193 |
22.34±0.091 |
1.05 |
0.8 |
|
BC333 |
66 |
21.32±0.075 |
24.66±0.041 |
21.1±0.158 |
-0.22 |
3.34 |
Table 2. PCR assay data for patients with diagnosed gastric cancer
All collumns as in Table 1.
|
Sample |
Age |
Sex |
I Cq_GGCC |
II Cq_CATG |
III Cq_A(5mC)GC |
ΔCq_T (II – I) |
ΔCq_5mC (III-I) |
|
GC38 |
60 |
f |
20.71±0.139 |
21.49±0.191 |
21.56±0.056 |
0.78 |
0.85 |
|
GC41 |
47 |
f |
22.08±0.102 |
27.23±0.25 |
21.93±0.193 |
5.15 |
-0.15 |
|
GC43 |
59 |
m |
22.24±0.071 |
23.31±0.023 |
23.02±0.055 |
1.07 |
0.78 |
|
GC45 |
51 |
m |
22.72±0.142 |
32.35±1.843 |
22.5±0.092 |
9.63 |
-0.22 |
|
GC47 |
62 |
m |
20.37±0.086 |
21.38±0.07 |
21.37±0.1 |
1.01 |
1 |
|
GC49 |
46 |
m |
21.28±0.081 |
28.17±0.139 |
21.38±0.068 |
6.89 |
0.1 |
|
GC50 |
53 |
m |
22.11±0.146 |
23.09±0.051 |
23.02±0.15 |
0.98 |
0.91 |
|
GC51 |
77 |
m |
21.1±0.133 |
28.24±0.361 |
20.96±0.056 |
7.14 |
-0.14 |
|
GC54 |
70 |
f |
20.62±0.057 |
21.85±0.07 |
21.54±0.133 |
1.23 |
0.92 |
|
GC80 |
72 |
m |
22.58±0.091 |
23.58±0.07 |
23.79±0.142 |
1 |
1.21 |
|
GC98 |
52 |
m |
21.33±0.068 |
22.23±0.077 |
22.16±0.013 |
0.9 |
0.83 |
|
GC99 |
56 |
m |
24.62±0.142 |
25.84±0.077 |
25.48±0.097 |
1.22 |
0.86 |
|
GC132 |
75 |
f |
21.08±0.028 |
28.96±0.596 |
20.92±0.152 |
7.88 |
-0.16 |
|
GC133 |
72 |
f |
21.88±0.141 |
22.94±0.076 |
22.69±0.168 |
1.06 |
0.81 |
|
GC146 |
80 |
m |
22.84±0.118 |
23.63±0.058 |
23.73±0.084 |
0.79 |
0.89 |
|
GC148 |
61 |
m |
21.27±0.06 |
28.37±0.325 |
21.37±0.165 |
7.1 |
0.1 |
|
GC149 |
83 |
m |
20.79±0.05 |
28.11±0.132 |
20.6±0.098 |
7.32 |
-0.19 |
|
GC150 |
57 |
m |
21.37±0.024 |
27.21±0.589 |
21.35±0.073 |
5.84 |
-0.02 |
|
GC151 |
50 |
m |
20.22±0.115 |
21.17±0.06 |
21.06±0.126 |
0.95 |
0.84 |
|
GC154 |
61 |
m |
20.95±0.05 |
22.07±0.05 |
21.9±0.218 |
1.12 |
0.95 |
|
GC156 |
70 |
m |
20.59±0.132 |
27.34±0.126 |
20.59±0.043 |
6.75 |
0 |
|
GC157 |
37 |
m |
21.91±0.144 |
23.1±0.116 |
22.87±0.124 |
1.19 |
0.96 |
|
GC 158 |
76 |
f |
22.16±0.044 |
30.49±1.099 |
22.17±0.177 |
8.33 |
0.01 |
|
GC161 |
49 |
m |
20.85±0.185 |
21.85±0.04 |
21.76±0.069 |
1 |
0.91 |
|
GC182 |
54 |
m |
20.21±0.054 |
21.31±0.036 |
21.26±0.116 |
1.1 |
1.05 |
|
GC184 |
69 |
m |
21.16±0.058 |
30.1±0.264 |
21.13±0.111 |
8.94 |
-0.03 |
|
GC185 |
69 |
m |
21.42±156 |
29.14±1.04 |
21.48±82 |
7.72 |
0.06 |
|
GC186 |
59 |
f |
21.19±0.029 |
21.95±0.181 |
22.07±0.165 |
0.88 |
0.76 |
|
GC187 |
70 |
f |
21.06±0.044 |
22.02±0.108 |
21.91±0.181 |
0.96 |
0.85 |
|
GC209 |
59 |
f |
21.06±0.02 |
21.82±0.163 |
22.05±0.175 |
0.99 |
0.76 |
|
GC211 |
65 |
f |
21.53±0.13 |
30.17±0.719 |
21.41±0.07 |
8.64 |
-0.12 |
|
GC214 |
82 |
f |
21.71±0.064 |
28.38±0.625 |
21.79±0.059 |
6.67 |
0.08 |
|
GC215 |
81 |
f |
21.34±0.086 |
22.22±0.026 |
22.22±0.128 |
0.88 |
0.88 |
|
GC220 |
48 |
m |
20.58±0.043 |
21.49±0.007 |
21.76±0.163 |
0.91 |
1.18 |
|
GC224 |
57 |
m |
21.57±0.136 |
29.9±0.476 |
21.48±0.148 |
8.33 |
-0.09 |
|
GC225 |
72 |
f |
21.59±0.096 |
29.69±0.964 |
21.58±0.162 |
8.1 |
-0.01 |
|
GC227 |
79 |
m |
21.5±0.081 |
29.63±0.38 |
21.47±0.089 |
8.13 |
-0.03 |
|
GC 230 |
63 |
f |
21.73±0.121 |
22.73±0.125 |
22.57±0.125 |
1 |
0.84 |
|
GC 232 |
64 |
f |
22.39±0.084 |
22.36±0.092 |
25.09±0.033 |
-0.03 |
2.7 |
|
GC 233 |
62 |
f |
21.17±0.048 |
22.17±0.064 |
21.93±0.077 |
1 |
0.76 |
|
GC 234 |
61 |
m |
21.21±0.039 |
22.13±0.047 |
22.15±0.107 |
0.94 |
0.92 |
|
GC243 |
75л |
m |
23.07±0.047 |
24.04±0.007 |
23.87±0.195 |
0.97 |
0.8 |
|
GC250 |
67 |
m |
22.73±0.163 |
29.5±0.41 |
22.56±0.049 |
6.77 |
-0.17 |
|
GC276 |
35 |
f |
21.23±0.058 |
26.34±0.982 |
21.29±0.107 |
5.11 |
0.06 |
|
GC277 |
72 |
f |
21.34±0.09 |
21.36±0.025 |
24.08±0.11 |
0.02 |
2.74 |
|
GC280 |
67 |
m |
22.34±0.122 |
23.4±0.031 |
23.11±0.069 |
1.06 |
0.77 |
|
GC282 |
68 |
f |
21.42±0.092 |
22.44±0.122 |
22.31±0.064 |
1.02 |
0.89 |
|
GC284 |
46 |
m |
22.56±0.065 |
23.74±0.186 |
23.35±0.076 |
1.18 |
0.79 |
|
GC285 |
71 |
f |
21.56±0.054 |
30.42±0.812 |
21.42±0.118 |
8.86 |
-0.14 |
|
GC286 |
66 |
m |
21.28±0.112 |
21.42±0.064 |
24.01±0.103 |
0.14 |
2.73 |
|
GC287 |
63 |
m |
21.17±0.128 |
22.18±0.072 |
22.06±0.023 |
1.01 |
0.89 |
|
GC315 |
74 |
f |
21.54±0.071 |
22.36±0.116 |
22.36±0.125 |
0.82 |
0.82 |
|
GC316 |
67 |
m |
20.54±0.146 |
21.75±0.049 |
21.38±0.075 |
1.21 |
0.84 |
|
GC 317 |
74 |
m |
21.62±0.037 |
28.98±0.159 |
21.47±0.022 |
7.36 |
-0.15 |
|
GC 318 |
71 |
m |
22.39±0.041 |
23.62±0.127 |
23.26±0.05 |
1.23 |
0.87 |
|
GC319 |
57 |
m |
21.51±0.046 |
29.65±0.212 |
21.64±0.139 |
8.14 |
0.13 |
|
GC334 |
67 |
f |
20.44±0.025 |
21.36±0.025 |
21.29±0.044 |
0.92 |
0.85 |
|
GC335 |
55 |
f |
21.52±0.025 |
21.35±0.2 |
24.21±0.035 |
-0.17 |
2.69 |
|
GC340 |
65 |
m |
22.21±0.018 |
22.13±0.129 |
24.2±0.176 |
-0.08 |
1.99 |
|
GC341 |
74 |
f |
22.45±0.08 |
23.29±0.135 |
23.34±0.09 |
0.84 |
0.89 |
|
GC342 |
72 |
m |
22.38±0.073 |
30.24±0.368 |
22.27±0.139 |
7.86 |
-0.11 |
|
GC343 |
72 |
m |
21.58±0.107 |
22.35±0.022 |
22.43±0.066 |
0.85 |
0.77 |
|
GC344 |
62 |
m |
22.23±0.085 |
30.9±1.269 |
22.27±0.107 |
8.67 |
0.04 |
Table 3.
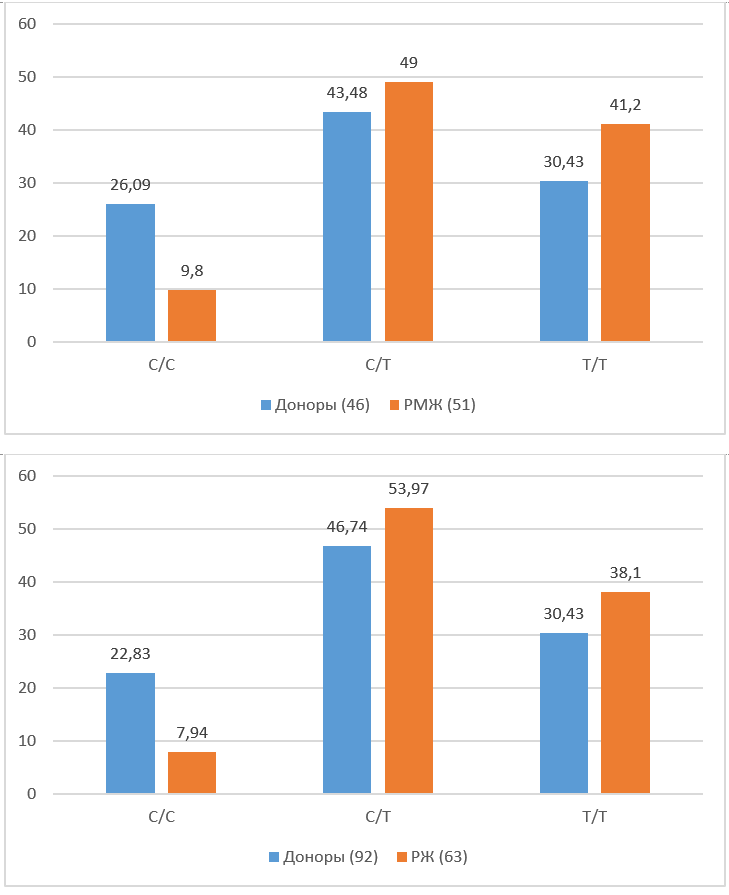
Distribution of diploid sets of alleles in position xp1: 245618129 in healthy people and patients with breast cancer and gastric cancer. The percentage values of the sample size are given; the number of people with a given diploid set is indicated in parentheses.
| Diploid sets | Healthy donors (46 ,
women) |
Breast cancer(51) | Healthy donors (92,
both sexes) |
Gastric cancer (63) |
| C/C | 26.09 (12) | 9.8 (5) | 22.83 (21) | 7.94 (5) |
| C/T | 43.48 (20) | 49 (25) | 46.74 (43) | 53.97 (34) |
| T/T | 30.43 (14) | 41.2 (21) | 30.43 (28) | 38.1 (24) |